Transcriptomics, also referred to as expression profiling, is the investigation of a set (or all) RNA molecules expressed in a certain tissue or cell type, in a specific developmental stage, or under certain physiological conditions. For us, transcriptomics plays an important role in the discovery of new candidate genes for functional studies, in addition to the creation of global expression profiles of individual cell types.
We are developing protocols to manually isolate living cells from flowering plant female and male gametophytes by micromanipulation with the aim to study the transcriptome of these cells. Previously, we used female gametes (egg cells and central cells) and two-celled proembryos of wheat to perform single-run partial sequencing of randomly selected cDNA clones (ESTs; “expressed sequence tags”) of generated cDNA libraries (Sprunck et al., 2005) to identify and investigate egg cell-specific and fertilization-induced gene products (Sprunck et al., 2012; Leljak-Levanić et al., 2013).
Meanwhile, we isolate plant gametes for next-generation sequencing (NGS) to investigate the mRNA and small RNA content of egg and sperm cells from different flowering plant species (e.g., Englhart et al., 2017; Chen et al., 2017; Flores-Tornero et al., 2019).
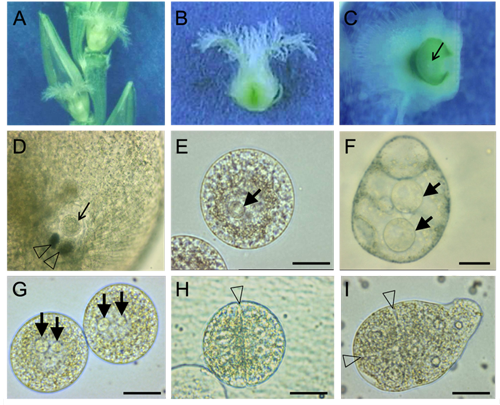
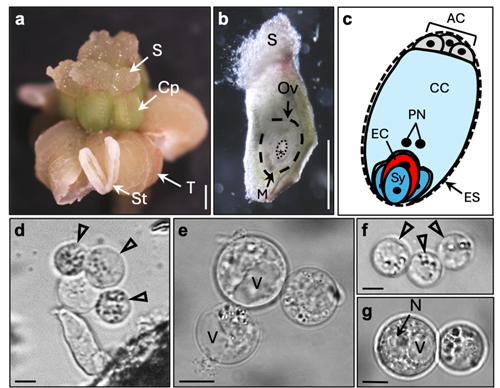
We are analyzing our transcriptome data sets with the aim to discover uncharacterized gamete-specific or fertilization-induced mRNAs, as well as small and long noncoding RNAs. We identified, for example, transcripts specifically present in the egg cell and/or the central cell and those up-regulated in zygotes or two-celled proembryos (Sprunck et al., 2005; Sprunck et al., 2012; Rademacher et al., 2013; Juranic et al., 2012; Leljak-Levanić et al., 2013; Chen et al., 2017; Bauer et al., 2019; Cyprys et al., 2019; Julca et al., 2021). Currently, we investigate the molecular function of selected candidate genes/ gene families/microRNAs in the establishment of cell polarity, in gamete specification, in gamete interactions during double fertilization, and in post-fertilization processes.
